7 Image From The Rscb Pdb Of Pdb Id Iznj Doi 10 2210 Pdb1znj Pdb
7 Image From The Rscb Pdb Of Pdb Id Iznj Doi 10 2210 Pdb1znj Pdb Protein secondary structure prediction (pssp) involves determining the local conformations of the peptide backbone in a folded protein, and is often the first step in resolving a protein’s global. Biological assembly 1 assigned by authors and generated by pisa (software).

Wwpdb 7jjj As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. the rcsb pdb also provides a variety of tools and resources. users can perform simple and advanced searches based on annotations relating to sequence, structure and function. these molecules are visualized, downloaded, and analyzed by users. The reported space group and unit cell parameters are almost identical to those of cubic insulin reported in the pdb. the results of x ray studies confirmed that the crystals obtained were cubic insulin crystals and that they contained no polysialic acid or its fragments. Create or download publication quality pictures of biomacromolecules with the rcsb pdb. several interactive, java based tools 1 can be used to visualize pdb data and create pictures. The protein data bank (pdb) the single global repository of experimentally determined 3d structures of biological macromolecules and their complexes was established in 1971, becoming the first open access digital resource in the biological sciences.
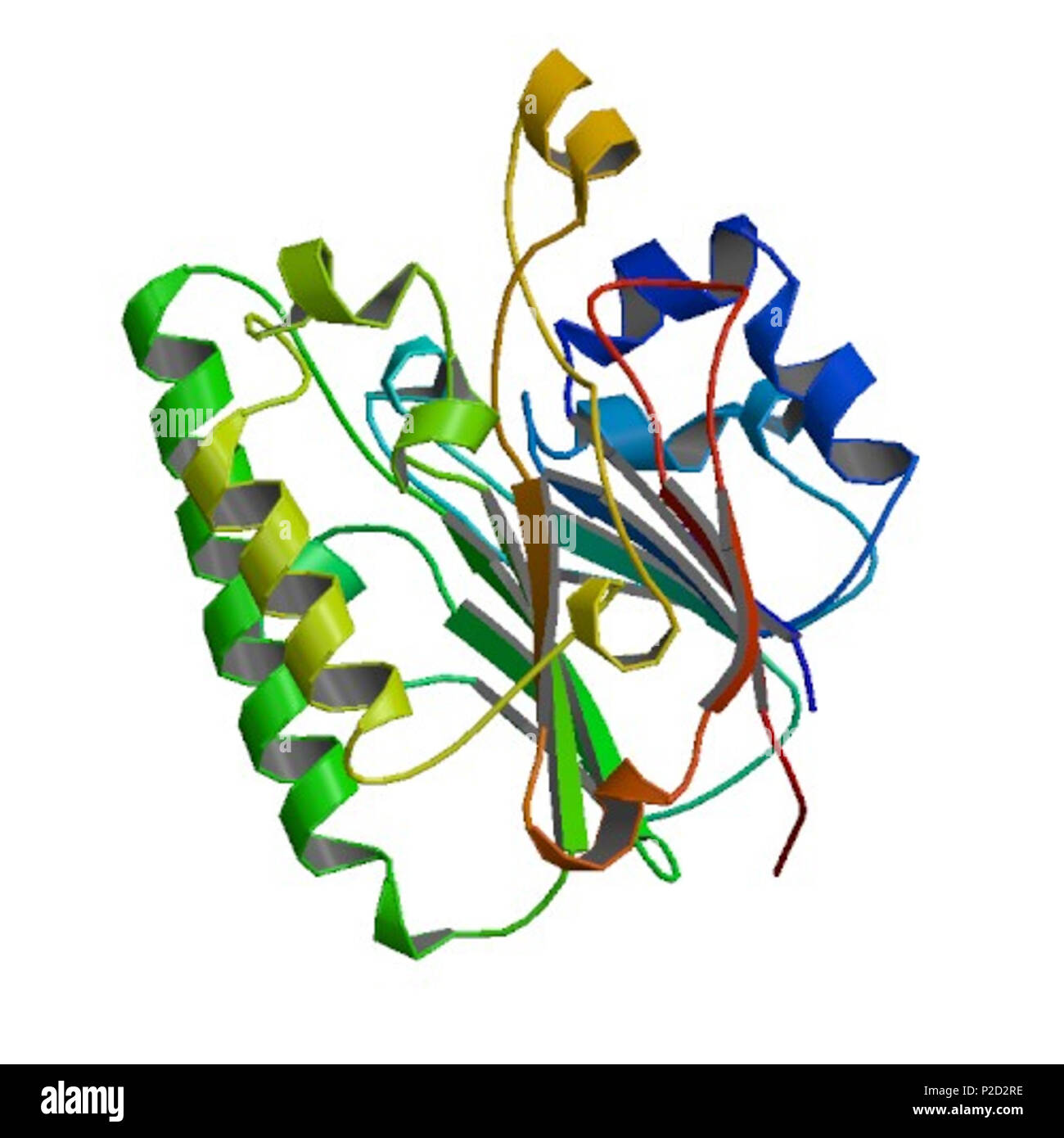
Imagesource Rcsb Pdb Structureid 4f1r Doi Http Dx Doi Org 10 2210 Create or download publication quality pictures of biomacromolecules with the rcsb pdb. several interactive, java based tools 1 can be used to visualize pdb data and create pictures. The protein data bank (pdb) the single global repository of experimentally determined 3d structures of biological macromolecules and their complexes was established in 1971, becoming the first open access digital resource in the biological sciences. Dois for pdb structures follow the format: 10.2210 pdbxxxx pdb, where xxxx is replaced with the pdb id (e.g., doi.org 10.2210 pdb4hhb pdb). doi citations should include the entry authors, deposition year, structure title, and doi. As a member of the wwpdb, the rcsb pdb curates and annotates pdb data according to agreed upon standards. the rcsb pdb also provides a variety of tools and resources. users can perform simple and advanced searches based on annotations relating to sequence, structure and function. The research collaboratory for structural bioinformatics protein data bank (rcsb pdb), founding member of the worldwide protein data bank (wwpdb), is the us data center for the open access pdb archive. A powerful new search system (search.rcsb.org) seamlessly integrates heterogeneous types of searches across the pdb archive. searches may combine text attributes, protein or nucleic acid sequences, small molecule chemical descriptors, 3d macromolecular shapes, and sequence motifs.
Comments are closed.