A Localization Of Potential Transcription Factor Binding Sites Of The

A Localization Of Potential Transcription Factor Binding Sites Of The These tfbss often localize near the transcriptional start site (tss) in an area termed the promoter, and specific locations elsewhere in the genome termed enhancers. these tfbss are bound by tfs that recruit additional proteins to either activate or repress gene expression. Given the limitations of previous research, we introduce an innovative deeputf model designed to model signals in transcription factor binding regions at the nucleotide level.

Localization Of Putative Stress Transcription Factor Binding Sites In Download scientific diagram | (a) localization of potential transcription factor binding sites of the transcription factors zkscan3, sox9 and hnf1α within the 100 ct fragment. Herein, we describe a robust approach that couples chromatin immunoprecipitation (chip) with the paired end ditag (pet) sequencing strategy for unbiased and precise global localization of tfbs. we have applied this strategy to map p53 targets in the human genome. This review summarizes approaches for in silico, in vitro, and in vivo identification of transcription factor binding sites. a variety of techniques useful for localized and high throughput analyses are discussed here, with emphasis on aspects of data generation and verification. To characterize transcription factor binding site function on a large scale, we predicted and mutagenized 455 binding sites in human promoters.

Illustrates The Position Of Potential Transcription Factor Binding This review summarizes approaches for in silico, in vitro, and in vivo identification of transcription factor binding sites. a variety of techniques useful for localized and high throughput analyses are discussed here, with emphasis on aspects of data generation and verification. To characterize transcription factor binding site function on a large scale, we predicted and mutagenized 455 binding sites in human promoters. We propose a integrative pipeline to explore the co localization of 55 tfs and 11 hms and its dynamics in human gm12878 and k562 by matched chip seq and rna seq data from encode. we classify tfs and hms into three types based on their binding enrichment around transcription start site (tss). Here, we use light controlled mutants of the yeast tf msn2 as a model system to investigate how promoter decoding of tf localization dynamics is affected by changes in the ability of the tf to bind dna. In this review, we discuss technological developments for identifying tf binding preferences and highlight recent discoveries that elaborate how tf interactions, local dna structure, and genomic features influence tf–dna binding. A major ques tion in current research is whether these potential binding sites are functional (for binding tfs, and for regulating transcription) and under what circumstances.

Models Of Transcription Factor Binding Sites Download Scientific Diagram We propose a integrative pipeline to explore the co localization of 55 tfs and 11 hms and its dynamics in human gm12878 and k562 by matched chip seq and rna seq data from encode. we classify tfs and hms into three types based on their binding enrichment around transcription start site (tss). Here, we use light controlled mutants of the yeast tf msn2 as a model system to investigate how promoter decoding of tf localization dynamics is affected by changes in the ability of the tf to bind dna. In this review, we discuss technological developments for identifying tf binding preferences and highlight recent discoveries that elaborate how tf interactions, local dna structure, and genomic features influence tf–dna binding. A major ques tion in current research is whether these potential binding sites are functional (for binding tfs, and for regulating transcription) and under what circumstances.
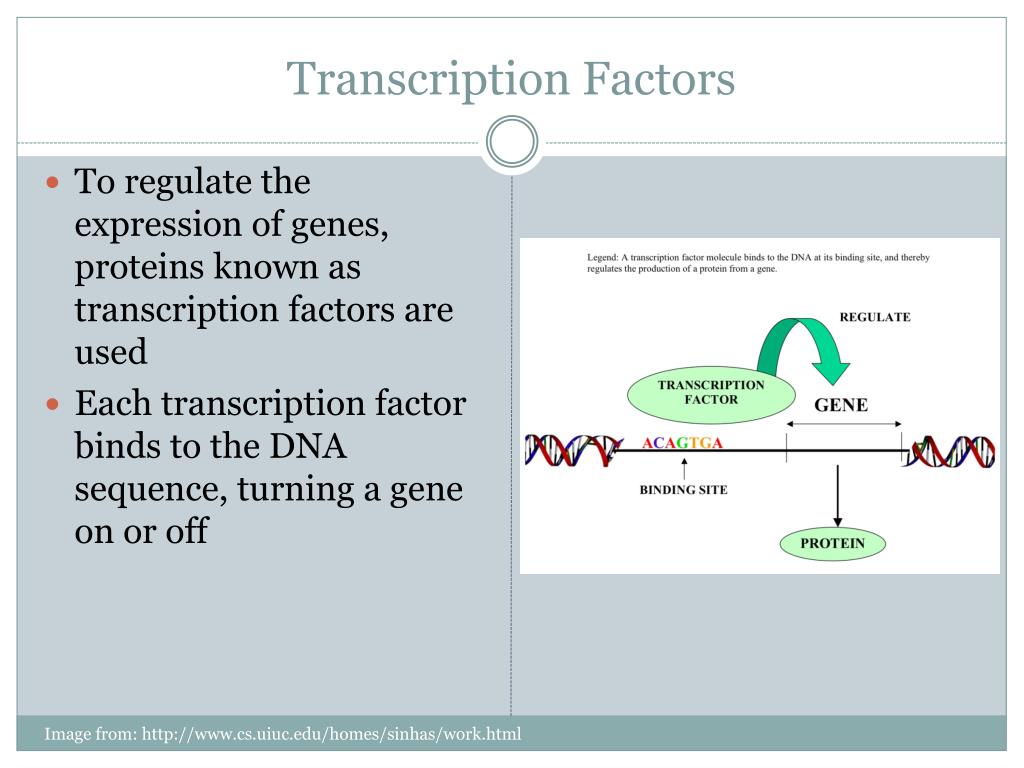
Ppt Detection Of Transcription Factor Binding Sites Powerpoint In this review, we discuss technological developments for identifying tf binding preferences and highlight recent discoveries that elaborate how tf interactions, local dna structure, and genomic features influence tf–dna binding. A major ques tion in current research is whether these potential binding sites are functional (for binding tfs, and for regulating transcription) and under what circumstances.

Identification Of Potential Transcription Factor Binding Sites In The
Comments are closed.