Comprehensive Workflow Of Mass Spectrometry Based Shotgun Proteomics Of

61786 T Png Here, we have described an optimized quantitative proteomics workflow for tissue samples using label free and label based proteome profiling methods, which is crucial for applications in life sciences, especially biomarker discovery based projects. “shotgun proteomics” or “bottom up proteomics” is the prevailing strategy, in which proteins are hydrolyzed into peptides that are analyzed by mass spectrometry.
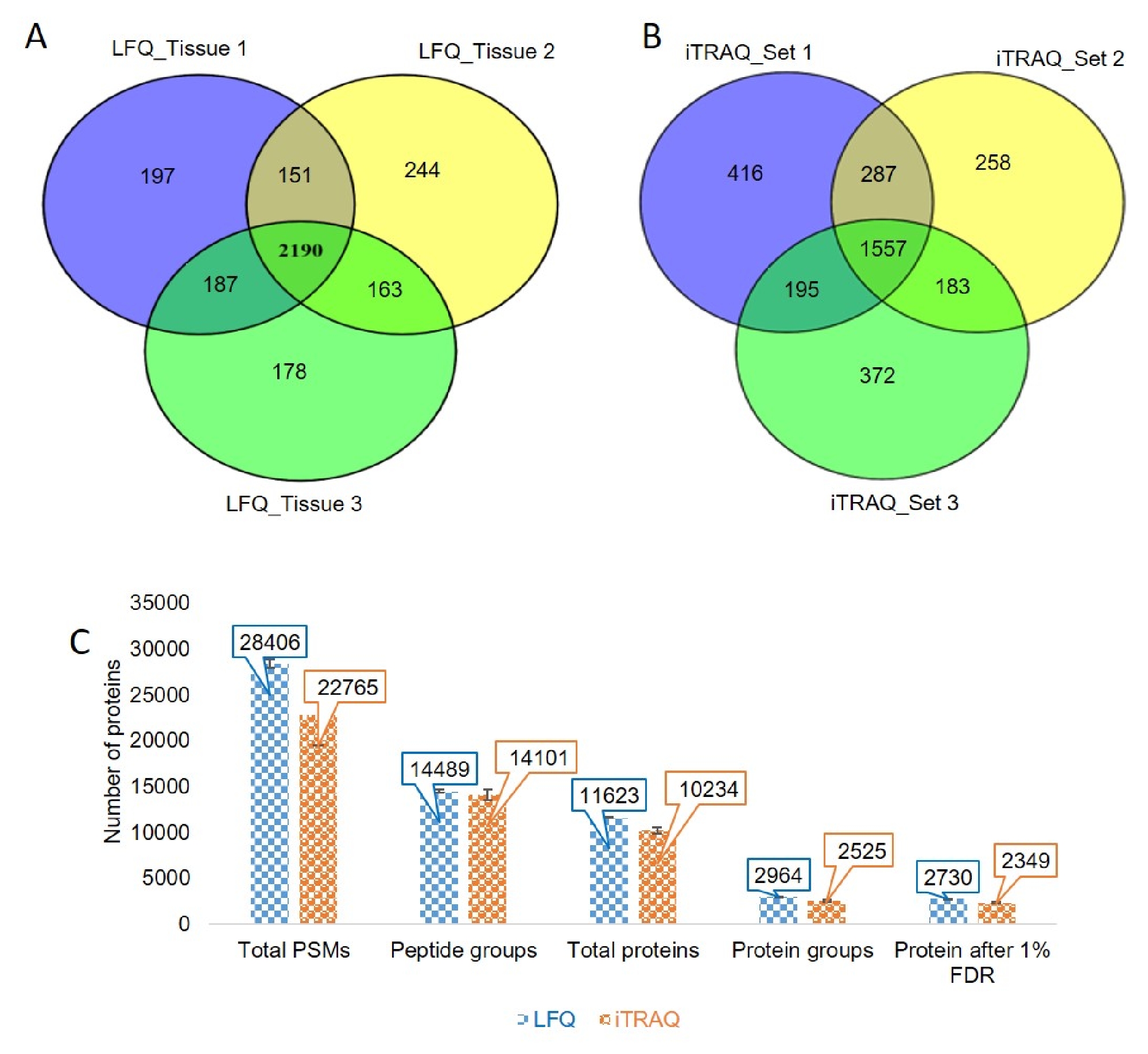
Comprehensive Workflow Of Mass Spectrometry Based Shotgun Proteomics Of Here we present an updated protocol covering the most important basic computational workflows, including those designed for quantitative label free proteomics, ms1 level labeling and isobaric. Here, we provide a comprehensive overview of different proteomics methods to aid the novice and experienced researcher. we cover from biochemistry basics and protein extraction to biological interpretation and orthogonal validation. This study investigates the challenge of comprehensively cataloging the complete human proteome from a single cell type using mass spectrometry (ms) based shotgun proteomics. This study investigates the challenge of comprehensively cataloging the complete human proteome from a single cell type using mass spectrometry (ms) based shotgun proteomics.
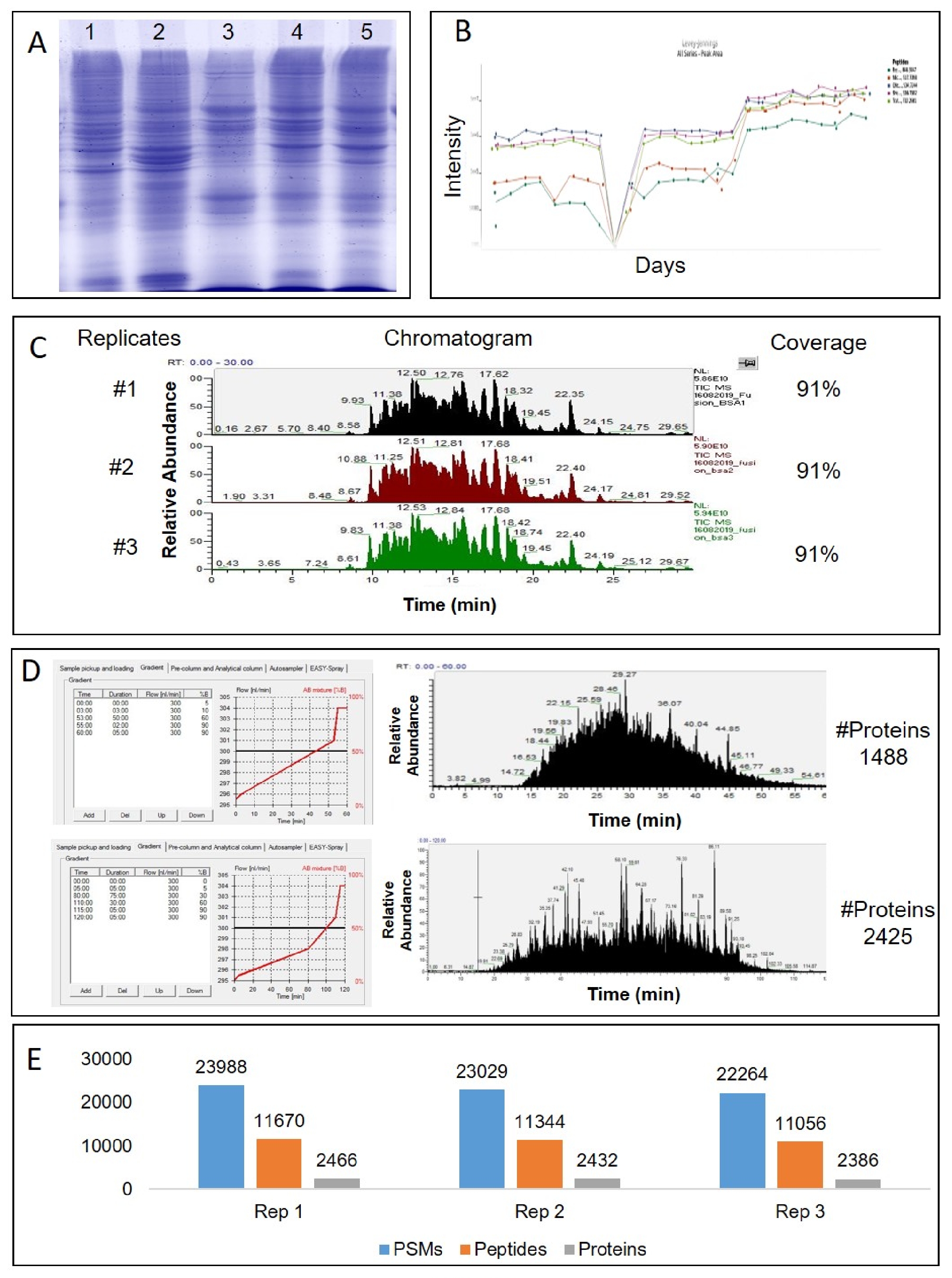
Comprehensive Workflow Of Mass Spectrometry Based Shotgun Proteomics Of This study investigates the challenge of comprehensively cataloging the complete human proteome from a single cell type using mass spectrometry (ms) based shotgun proteomics. This study investigates the challenge of comprehensively cataloging the complete human proteome from a single cell type using mass spectrometry (ms) based shotgun proteomics. Maxquant is one of the most frequently used platforms for mass spectrometry (ms) based proteomics data analysis. since its first release in 2008, it has grown substantially in functionality. Over the past 2 to 3 years, mass spectrometry based single cell proteomics (scp) has experienced transformative improvements in microfluidic and robotic sample preparation, innovative ms1 and ms2 based multiplexing strategies, and specialized hardware (e.g., timstof ultra 2, astral), which have dramatically boosted sensitivity, throughput, and proteome coverage from picogram level protein. During the past decade, various computational methods have been developed to integrate orthogonal data sources to improve peptide and protein identification in shotgun proteomics studies. these approaches take advantage of the rapidly growing volumes of genomic, transcriptomic, and interactome data. Shotgun (or bottom up) proteomics using mass spectrometry has become the most widely used method for unbiased protein quantification from biological samples. major progress over the last 30 years has enabled over 10,000 proteins to be identified and quantified from a single analysis [1].

Comprehensive Workflow Of Mass Spectrometry Based Shotgun Proteomics Of Maxquant is one of the most frequently used platforms for mass spectrometry (ms) based proteomics data analysis. since its first release in 2008, it has grown substantially in functionality. Over the past 2 to 3 years, mass spectrometry based single cell proteomics (scp) has experienced transformative improvements in microfluidic and robotic sample preparation, innovative ms1 and ms2 based multiplexing strategies, and specialized hardware (e.g., timstof ultra 2, astral), which have dramatically boosted sensitivity, throughput, and proteome coverage from picogram level protein. During the past decade, various computational methods have been developed to integrate orthogonal data sources to improve peptide and protein identification in shotgun proteomics studies. these approaches take advantage of the rapidly growing volumes of genomic, transcriptomic, and interactome data. Shotgun (or bottom up) proteomics using mass spectrometry has become the most widely used method for unbiased protein quantification from biological samples. major progress over the last 30 years has enabled over 10,000 proteins to be identified and quantified from a single analysis [1].

Comprehensive Workflow Of Mass Spectrometry Based Shotgun Proteomics Of During the past decade, various computational methods have been developed to integrate orthogonal data sources to improve peptide and protein identification in shotgun proteomics studies. these approaches take advantage of the rapidly growing volumes of genomic, transcriptomic, and interactome data. Shotgun (or bottom up) proteomics using mass spectrometry has become the most widely used method for unbiased protein quantification from biological samples. major progress over the last 30 years has enabled over 10,000 proteins to be identified and quantified from a single analysis [1].
Comments are closed.