Exploring Pdb Structures In 3d With Molstar Mol Introductory Guide

Mol Viewer Pdf Protein Data Bank Biology This tutorial series is an introductory guide to visualizing pdb structures and creating graphics and animations with the mol* viewer available at www. The tutorial series below is an introductory guide to visualizing pdb structures and creating graphics and animations with the mol* viewer. to access the individual sections, click on the links under each tutorial and hit “play” on the video.

Pdb 101 Learn Videos Exploring Pdb Structures In 3d With Molstar It adopts declarative data driven approach to describe, load, render, and visually deliver molecular structures, along with 3d representations, coloring schemes, and associated structural, biological, or functional annotations. Select all or parts of the 3d structure to display it in different representations, colors, and more, to analyze interactions, and to measure distances and angles within and between proteins, nucleic acids, and other molecules. Mol* is used throughout rcsb.org, from generating the 2d images available for every structure and entity in the pdb; to offering interactive 3d views of structures, sequence annotations on structures, electron density; and close up views of ligand interactions. The participants had the opportunity to learn about the protein structure related tools, visualizations, and workflows that have been integrated into doe kbase.

Pdb 101 Learn Videos Exploring Pdb Structures In 3d With Molstar Mol* is used throughout rcsb.org, from generating the 2d images available for every structure and entity in the pdb; to offering interactive 3d views of structures, sequence annotations on structures, electron density; and close up views of ligand interactions. The participants had the opportunity to learn about the protein structure related tools, visualizations, and workflows that have been integrated into doe kbase. There are several ways to make selections in mol*. you first need to open selection mode and change the picking level, if needed. to make selections, you can either click on parts of the structure in the 3d canvas or sequence panel, or use the set operations menus in the selection mode toolbar. Mol* is used throughout rcsb.org, from generating the 2d images available for every structure and entity in the pdb; to offering interactive 3d views of structures, sequence annotations on structures, electron density; and close up views of ligand interactions. Mol* ("molstar") is a molecular visualization program that can be used to import, visualize, and align multiple structures from the protein data bank (pdb). it is the main 3d. All the interactions with the molecule (s) uploaded in mol* require using mouse controls (in the 3d canvas). these controls allow the user to manipulate the view of structures via a variety of functions such as rotating, translating, zooming, and clipping the structures.
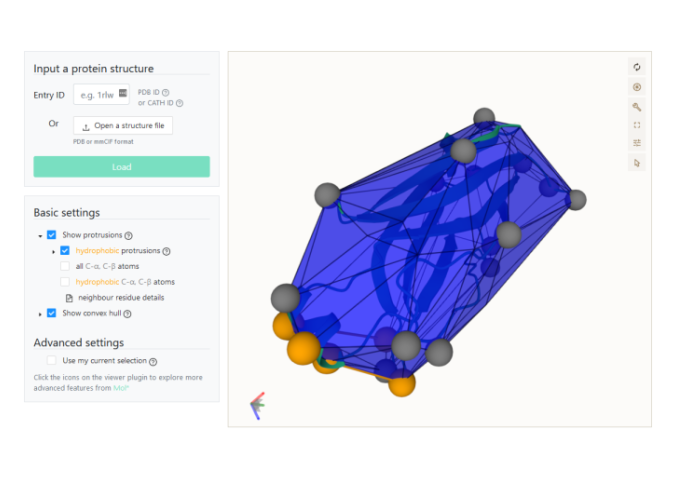
Mol There are several ways to make selections in mol*. you first need to open selection mode and change the picking level, if needed. to make selections, you can either click on parts of the structure in the 3d canvas or sequence panel, or use the set operations menus in the selection mode toolbar. Mol* is used throughout rcsb.org, from generating the 2d images available for every structure and entity in the pdb; to offering interactive 3d views of structures, sequence annotations on structures, electron density; and close up views of ligand interactions. Mol* ("molstar") is a molecular visualization program that can be used to import, visualize, and align multiple structures from the protein data bank (pdb). it is the main 3d. All the interactions with the molecule (s) uploaded in mol* require using mouse controls (in the 3d canvas). these controls allow the user to manipulate the view of structures via a variety of functions such as rotating, translating, zooming, and clipping the structures.
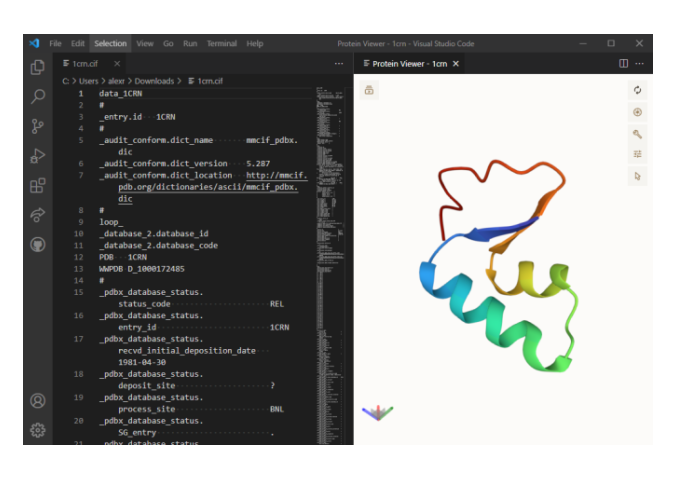
Mol Mol* ("molstar") is a molecular visualization program that can be used to import, visualize, and align multiple structures from the protein data bank (pdb). it is the main 3d. All the interactions with the molecule (s) uploaded in mol* require using mouse controls (in the 3d canvas). these controls allow the user to manipulate the view of structures via a variety of functions such as rotating, translating, zooming, and clipping the structures.
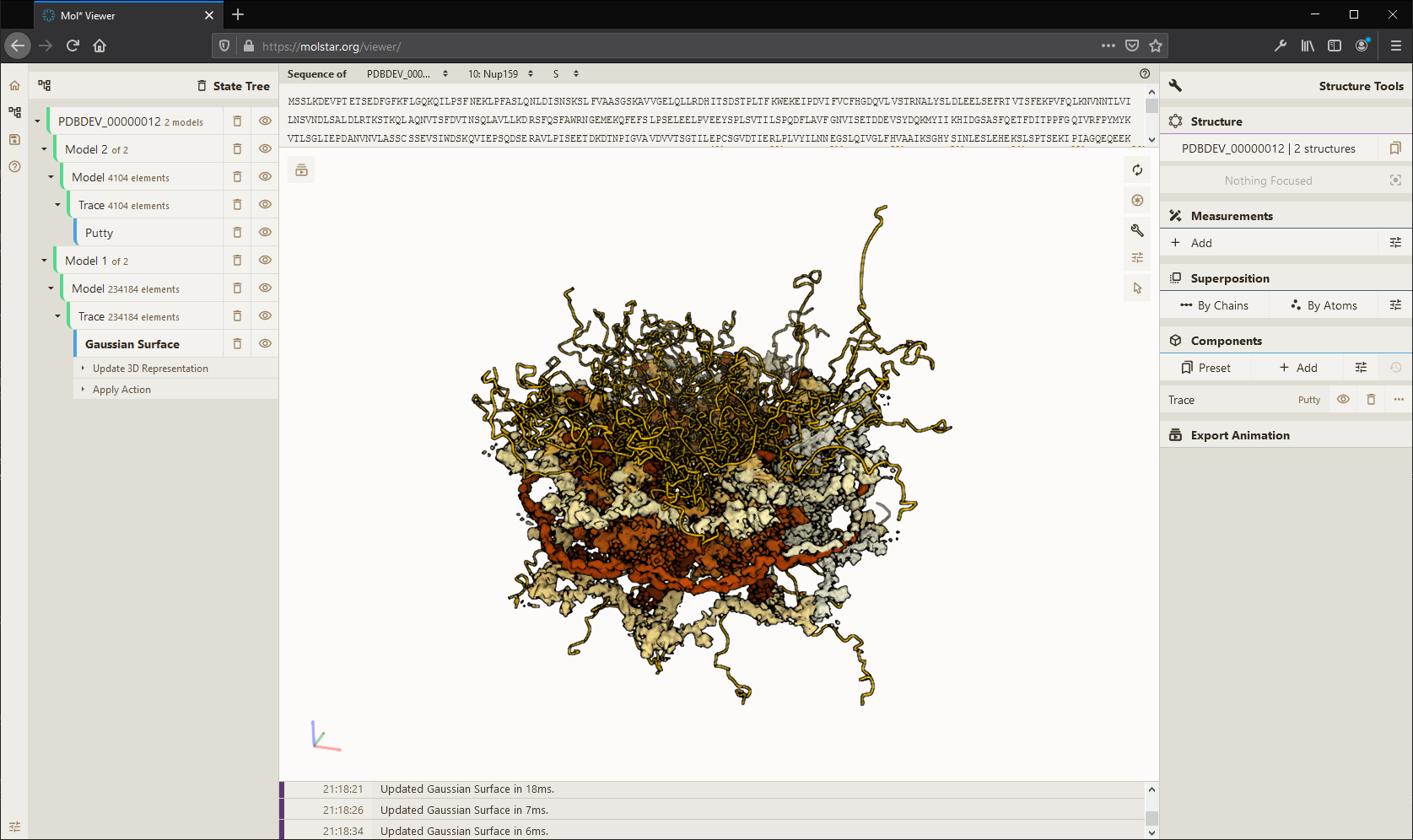
Mol
Comments are closed.