Using Single Cell And Spatial Transcriptomics To Understand Tumors
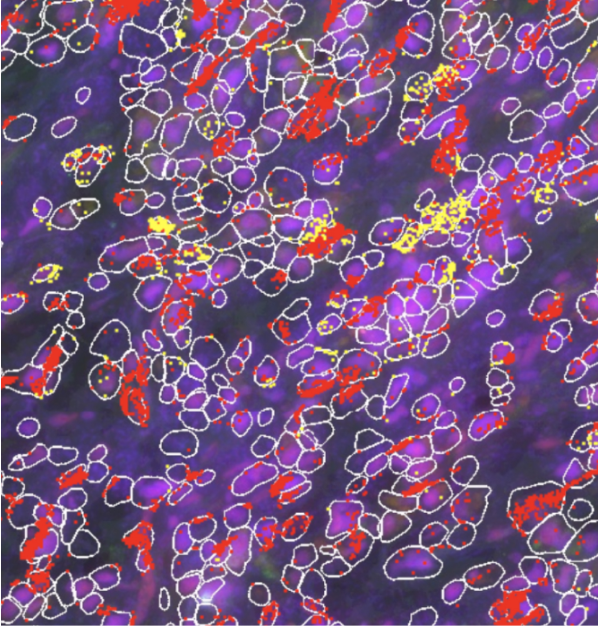
Talk2data Single Cell And Spatial Analytics Hub Here, we use single cell, spatial and in situ technologies on serial sections of an ffpe preserved breast cancer block to explore the heterogeneity within the tumor (fig. 1). This review integrates recent advancements in single cell rna sequencing (scrna seq) and spatial transcriptomics to elucidate glioblastoma’s cellular heterogeneity and metabolic reprogramming.

The Full Diagram Of Single Cell Transcriptomics Data Capture Gene Single cell and spatial transcriptomics sequencing have recently emerged as key technologies for unravelling tumour heterogeneity. single cell sequencing promotes individual cell type identification through transcriptome wide gene expression measurements of each cell. Chen et al. integrate sc snrna seq with spatial transcriptomics to characterize cellular composition, lineage dynamics, and spatial architecture in pdac tissues with varying neural invasion status, uncovering distinct peri and intra neural microenvironments and identifying tgfbi schwann cells as promoters of neural invasion. Comprehensive spatial analysis of the tumor microenvironment (tme) of cancer research samples has huge potential to reveal the cellular and molecular mechanisms of disease or resistance and, ultimately, point to putative drug targets for therapeutic development. To understand the crosstalk between diverse cellular components in proximity in the tme, we performed scrna seq coupled with spatial transcriptomic (st) assay to profile 41,700 cells from.

Using Single Cell And Spatial Transcriptomics To Dissect Cellular Comprehensive spatial analysis of the tumor microenvironment (tme) of cancer research samples has huge potential to reveal the cellular and molecular mechanisms of disease or resistance and, ultimately, point to putative drug targets for therapeutic development. To understand the crosstalk between diverse cellular components in proximity in the tme, we performed scrna seq coupled with spatial transcriptomic (st) assay to profile 41,700 cells from. Single cell rna seq technique and spatial transcriptomics are valuable platforms for the identification of cancer cells in their physical locations, which can be used for exploring intra tumor heterogeneity and epigenetic modifications for individualized therapeutic approaches. We asked experts to discuss some aspects of this technology from revealing the tumor microenvironment and heterogeneity, to tracking tumor evolution, to guiding tumor therapy, to current technical challenges. Introduction recent advancements in transcriptomic analysis, combined with single cell rna sequencing (scrna seq) and spatial transcriptomics, have deepened our understanding of the tumor microenvironment and cellular heterogeneity, laying the groundwork for personalized therapies. the aim of this research is to explore the heterogeneity of tumor cells in colorectal cancer (crc) and evaluate. Here, we propose scmalignantfinder, a machine learning tool specifically designed to distinguish malignant cells from their normal counterparts using a data and knowledge driven strategy.

Mapping Tumors Genes Spatial Transcriptomics Man Cave Health Single cell rna seq technique and spatial transcriptomics are valuable platforms for the identification of cancer cells in their physical locations, which can be used for exploring intra tumor heterogeneity and epigenetic modifications for individualized therapeutic approaches. We asked experts to discuss some aspects of this technology from revealing the tumor microenvironment and heterogeneity, to tracking tumor evolution, to guiding tumor therapy, to current technical challenges. Introduction recent advancements in transcriptomic analysis, combined with single cell rna sequencing (scrna seq) and spatial transcriptomics, have deepened our understanding of the tumor microenvironment and cellular heterogeneity, laying the groundwork for personalized therapies. the aim of this research is to explore the heterogeneity of tumor cells in colorectal cancer (crc) and evaluate. Here, we propose scmalignantfinder, a machine learning tool specifically designed to distinguish malignant cells from their normal counterparts using a data and knowledge driven strategy.
Comments are closed.